Discovery Genomics services include QC of all submitted samples prior to processing. If samples fail our QC metrics you will be contacted. You may choose to send replacement samples or proceed with the failed samples. Please note that we cannot guarantee successful library preparation or sequencing results for samples that have failed our QC metrics.
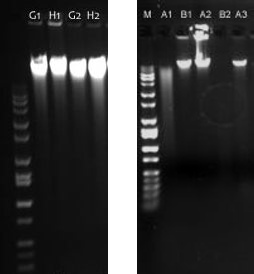
Visualization of gDNA by gel. The gel image on the left shows high-quality, high-molecular weight, intact gDNA in all wells. The gel image on right shows several samples that would not pass Discovery Genomics QC. Sample A1 shows a smear of gDNA indicating degradation. Sample A2 also shows intact gDNA, but as evidenced by the excess sample remaining in the well, the sample was provided at a concentration much higher than requested. Sample B2 shows no intact gDNA. Samples B1 and A3 show high-quality gDNA that would perform well in Discovery Genomics protocols.
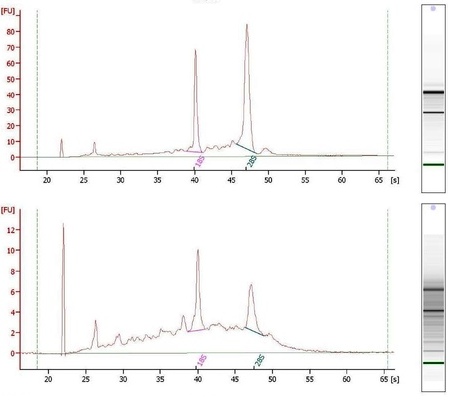
Agilent 2100 Bioanalyzer electropherograms for two RNA samples.
The electropherogram on top shows good-quality RNA with a 28S/18S rRNA ratio of 1.7 and an
RNA Integrity Number of 8.9 (max is 10.0). Note the rRNA peaks are distinct, with 28S higher than the
18S, and there are only a few smaller peaks that indicate RNA degradation. This sample should perform
well in Discovery Genomics protocols.
The electropherogram on the bottom shows poor-quality RNA with a RIN of 6.
The 28S rRNA peak is shorter than the 18S peak and many smaller peaks are seen along the baseline,
indicating a fair amount of RNA degradation. This sample would not pass Discovery Genomics QC requirements for use in
most protocols.
Discovery Genomics services include QC of all submitted samples prior to processing. Some quality measures such as RIN and 28S to 18S ratio are based on certain species (human, mouse, rat) and other organisms’ values for those metrics may differ. For example, insect bioanalysis profiles may not generate good RIN numbers, but still be high quality, intact RNA that will perform well in protocols. You should find the expected profile for your species and assess the quality by comparing our bioanalysis results to that profile. When an RNA profile is atypical, we require your approval before continuing with the sample. If samples fail our QC metrics you will be contacted. You may choose to send replacement samples or proceed with the failed samples. Please note that we cannot guarantee successful library preparation or sequencing results for samples that have failed our QC metrics.